😎 공부하는 징징알파카는 처음이지?
[Kaggle] Chest X-Ray 폐암 이미지 분류하기 본문
728x90
반응형
220129 작성
<본 블로그는 Kaggle 을 참고해서 공부하며 작성하였습니다>
Search | Kaggle
www.kaggle.com
폐암 이미지 분류하기
캐글의 Chest X-Ray Images 사용폐렴은 폐에 염증이 생긴 상태로 중증의 호흡기 감염병입니다.아래 이미지처럼 폐부위에 희미한 그림자?같은게 보이는데 사실 이 사진만으로 확실히 폐렴이다 아니다
velog.io
1. 라이브러리 import 및 데이터 load
import os, re
import random, math
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt# 데이터 로드 빠르게
AUTOTUNE = tf.data.experimental.AUTOTUNE
# X-ray 이미지 사이즈
IMAGE_SIZE = [180, 180]- 데이터 경로
root_path = os.getcwd() + '\\'
train_path = root_path + "chest_xray\\train\\*\\*"
val_path = root_path + "chest_xray\\val\\*\\*"
test_path = root_path + "chest_xray\\test\\*\\*"BATCH_SIZE = 30 # 정한 만큼의 배치
EPOCHS = 50
2. 데이터 준비
- train_data : 5216
- test_data : 624
- val_data : 16
train_data = tf.io.gfile.glob(train_path)
test_data = tf.io.gfile.glob(test_path)
val_data = tf.io.gfile.glob(val_path)
print(len(train_data))
print(len(test_data))
print(len(val_data))-
val 너무 작으니 train 과 val 합치고 다시 8 : 2
train_val_sum = tf.io.gfile.glob(train_path)
train_val_sum.extend(tf.io.gfile.glob(val_path))
# 8:2
train_size = math.floor(len(train_val_sum)*0.8)
random.shuffle(train_val_sum) # shuffle : 고정 크기 버퍼를 유지하고 뮤작위로 균일
train = train_val_sum[:train_size]
val = train_val_sum[train_size:]
print(len(train))
print(len(val))- train : 4185
- val : 1047
- 폐렴 아닌 normal : 1077
- 폐렴 : 3108
normal = len([filename for filename in train if "NORMAL" in filename])
print(f"normal image count in train set : {normal}")
pneumonia = len([filename for filename in train if "PNEUMONIA" in filename])
print(f"pneumonia image count in train set : {pneumonia}")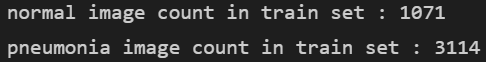
-
tf.data.Dataset.from_tensor_slices 함수는 tf.data.Dataset 를 생성하는 함수
train_list_ds = tf.data.Dataset.from_tensor_slices(train)
val_list_ds = tf.data.Dataset.from_tensor_slices(val)TRAIN_IMG_COUNT = tf.data.experimental.cardinality(train_list_ds).numpy()
print(f"Training images count: {TRAIN_IMG_COUNT}")
VAL_IMG_COUNT = tf.data.experimental.cardinality(val_list_ds).numpy()
print(f"Validating images count: {VAL_IMG_COUNT}")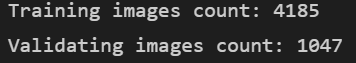
-
'NORMAL'이나 'PNEUMONIA'가 포함되어 있기 때문에 이를 이용해서 라벨 데이터
# 파일 경로의 끝에서 두번째 부분으로 양성과 음성을 구분
def get_label(file_path):
parts = tf.strings.split(file_path, os.path.sep)
return parts[-2] == "PNEUMONIA" # 폐렴이면 양성(True), 노말이면 음성(False)
- 이미지 사이즈 줄이기
def decode_img(img):
img = tf.image.decode_jpeg(img, channels=3) # 이미지를 uint8 tensor로 수정
img = tf.image.convert_image_dtype(img, tf.float32) # float32 타입으로 수정
img = tf.image.resize(img, IMAGE_SIZE) # 이미지 사이즈를 [180, 180] 수정
return img- 이미지, 라벨 읽기
def process_path(file_path):
label = get_label(file_path) # 라벨 검출
img = tf.io.read_file(file_path) # 이미지 읽기
img = decode_img(img) # 이미지를 알맞은 형식으로 수정
return img, label
- train, validation 데이터 셋 만들기
# 빠르게 데이터 처리
train_ds = train_list_ds.map(process_path, num_parallel_calls=AUTOTUNE)
val_ds = val_list_ds.map(process_path, num_parallel_calls=AUTOTUNE)
- 이미지 확인
for image, label in train_ds.take(1):
print("Image shape: ", image.numpy().shape)
print("Label: ", label.numpy())
- test 도 동일하게
test_list_ds = tf.data.Dataset.list_files(test_path)
TEST_IMAGE_COUNT = tf.data.experimental.cardinality(test_list_ds).numpy()
test_ds = test_list_ds.map(process_path, num_parallel_calls=AUTOTUNE)
test_ds = test_ds.batch(BATCH_SIZE)
for image, label in test_ds.take(1):
print("Image shape: ", image.numpy().shape)
print("Label: ", label.numpy())
print(TEST_IMAGE_COUNT)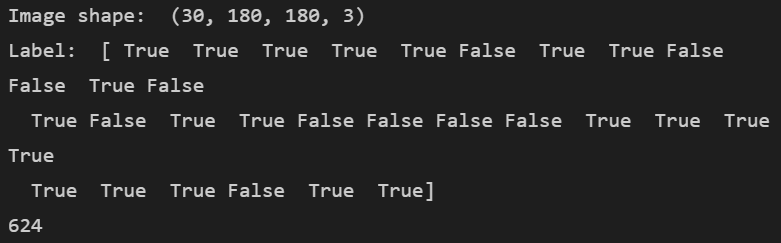
- data 를 좀더 효율적으로
# random_flip_left_right : 랜덤하게 좌우를 반전
def augment(image,label):
image = tf.image.random_flip_left_right(image)
return image,labeldef prepare_for_training(ds, shuffle_buffer_size=1000):
ds = ds.map(
augment, # augment 함수 적용
num_parallel_calls=2
)
# shuffle : 고정 크기 버퍼를 유지하고 뮤작위로 균일하게 다음 요소를 선택
ds = ds.shuffle(buffer_size=shuffle_buffer_size)
# repeat : 여러 번 데이터 부르기
ds = ds.repeat()
# batch : 정한 만큼의 배치
ds = ds.batch(BATCH_SIZE)
# prefech : GPU와 CPU를 효율적으로 사용
ds = ds.prefetch(buffer_size=AUTOTUNE)
return dstrain_ds = prepare_for_training(train_ds)
val_ds = prepare_for_training(val_ds)
3. 데이터 시각화
# 이미지 배치를 입력하면 여러장의 이미지를 보여줌
def show_batch(image_batch, label_batch):
plt.figure(figsize=(10,10))
for n in range(BATCH_SIZE): # BATCH_SIZE = 30
ax = plt.subplot(5,math.ceil(BATCH_SIZE / 5),n+1)
plt.imshow(image_batch[n])
if label_batch[n]:
plt.title("PNEUMONIA")
else:
plt.title("NORMAL")
plt.axis("off")image_batch, label_batch = next(iter(train_ds))
show_batch(image_batch.numpy(), label_batch.numpy())
4. CNN 모델링
: CNN은 Convolution Layer와 Max Pooling 레이어를 반복적으로 stack을 쌓는 특징 추출(Feature Extraction)
: Fully Connected Layer를 구성하고 마지막 출력층에 Softmax를 적용한 분류 부분으로 나뉜다
- Convolution Block
1) conv 2 번
2) batch normal 로 과적합 방지
3) max pooling
def conv_block(filters):
block = tf.keras.Sequential([
tf.keras.layers.SeparableConv2D(filters, 3, activation='relu', padding='same'),
tf.keras.layers.SeparableConv2D(filters, 3, activation='relu', padding='same'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.MaxPool2D()
])
return block- dense block
def dense_block(units, dropout_rate):
block = tf.keras.Sequential([
tf.keras.layers.Dense(units, activation='relu'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(dropout_rate)
])
return block- 전체 모델
def build_model():
model = tf.keras.Sequential([
tf.keras.layers.InputLayer(shape=(IMAGE_SIZE[0], IMAGE_SIZE[1], 3)),
tf.keras.layers.Conv2D(16, 3, activation='relu', padding='same'),
tf.keras.layers.Conv2D(16, 3, activation='relu', padding='same'),
tf.keras.layers.MaxPool2D(),
conv_block(32),
conv_block(64),
conv_block(128),
tf.keras.layers.Dropout(0.2),
conv_block(256),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Flatten(),
dense_block(512, 0.7),
dense_block(128, 0.5),
dense_block(64, 0.3),
tf.keras.layers.Dense(1, activation='sigmoid')
])
return model
5. 데이터 불균형 처리
- 랜덤 포레스트가 좋다
weight_for_0 = (1 / normal)*(TRAIN_IMG_COUNT)/2.0
weight_for_1 = (1 / pneumonia)*(TRAIN_IMG_COUNT)/2.0
class_weight = {0: weight_for_0, 1: weight_for_1}
print('Weight for NORMAL: {:.2f}'.format(weight_for_0))
print('Weight for PNEUMONIA: {:.2f}'.format(weight_for_1))
6. 모델 훈련
with tf.device('/GPU:0'):
model = build_model()
METRICS = [
'accuracy',
tf.keras.metrics.Precision(name='precision'),
tf.keras.metrics.Recall(name='recall')
]
model.compile(
optimizer='adam',
loss='binary_crossentropy',
metrics=METRICS
)- EarlyStopping
from keras.callbacks import EarlyStopping
# 최고의 정확도를 가질때 멈춰주는 함수 추가
es=EarlyStopping(monitor='val_loss',mode='min',verbose=1,patience=10)history = model.fit(
train_ds,
steps_per_epoch = TRAIN_IMG_COUNT // BATCH_SIZE,
epochs=10, callbacks=[es],
validation_data = val_ds,
validation_steps = VAL_IMG_COUNT // BATCH_SIZE,
class_weight = class_weight,
)
7. 시각화
fig, ax = plt.subplots(1, 4, figsize=(20, 5))
ax = ax.ravel()
for i, met in enumerate(['precision', 'recall', 'accuracy', 'loss']):
ax[i].plot(history.history[met])
ax[i].plot(history.history['val_' + met])
ax[i].set_title('Model {}'.format(met))
ax[i].set_xlabel('epochs')
ax[i].set_ylabel(met)
ax[i].legend(['train', 'val'])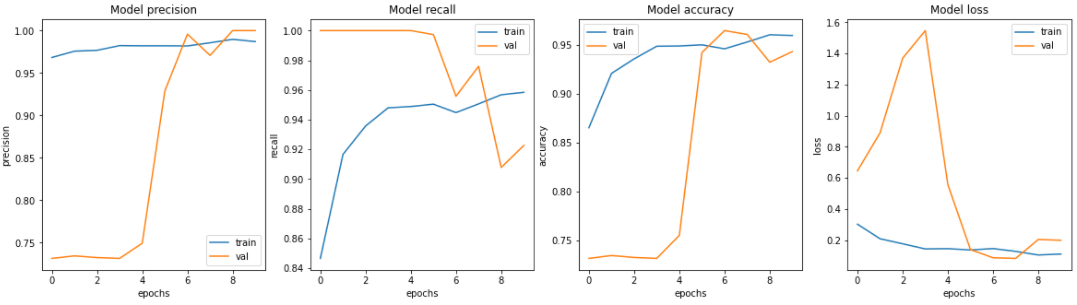
- test 데이터로 모델 평가
from IPython.display import Image
from IPython.core.display import HTML
Image(url= "https://miro.medium.com/proxy/1*pOtBHai4jFd-ujaNXPilRg.png")
Precision ( 정밀도 ) ex) 확진자로 분류된 사람들 중 실제 양성 시미닐 확률
Recall ( 재현율 ) ex) 실제로 양성인 시민을 확진자로 분류할 확률
loss, accuracy, precision, recall = model.evaluate(test_ds)
print(f'Loss: {loss},\nAccuracy: {accuracy},\nPrecision: {precision},\nRecall: {recall}')- Precision 과 Recall 은 trade-off 관계

728x90
반응형
'👩💻 컴퓨터 구조 > Kaggle' 카테고리의 다른 글
| [Kaggle]Super Image Resolution_고화질 이미지 만들기 (0) | 2022.02.07 |
|---|---|
| [Kaggle] CNN Architectures (0) | 2022.02.04 |
| [Kaggle] HeartAttack 예측 (0) | 2022.01.31 |
| [Kaggle]Breast Cancer Wisconsin (Diagnostic) Data Set_유방암 분류 (0) | 2022.01.28 |
| [Kaggle]MBTI_Myers-Briggs Personality Type Dataset(성격연구) (0) | 2022.01.22 |
Comments




