😎 공부하는 징징알파카는 처음이지?
[DACON] HAICon2020 산업제어시스템 보안위협 탐지 AI & LSTM 본문
728x90
반응형
220907 작성
<본 블로그는 dacon 대회에서의 Sllab 팀 코드와 dacon의 HAI 2.0 Baseline 글을 참고해서 공부하며 작성하였습니다 :-) >
https://dacon.io/competitions/official/235624/codeshare/1570?page=1&dtype=recent
HAI 2.0 Baseline
HAICon2020 산업제어시스템 보안위협 탐지 AI 경진대회
dacon.io
https://dacon.io/competitions/official/235624/codeshare/1831
[2위]SIlab
HAICon2020 산업제어시스템 보안위협 탐지 AI 경진대회
dacon.io
😎 1. 대회 소개
- 최근 국가기반시설 및 산업시설의 제어시스템에 대한 사이버 보안위협이 지속적으로 증가
- 현장 제어시스템의 특성을 정확하게 반영하고, 다양한 제어시스템 사이버공격 유형을 포함하는 데이터셋은 AI기반 보안기술 연구를 위한 필수적인 요소
- 산업제어시스템 보안 데이터셋 HAI 1.0 (제어시스템 테스트베드) 을 활용하여 정상 상황의 데이터만을 학습하여 공격 및 비정상 상황을 탐지할 수 있는 최신의 머신러닝, 딥러닝 모델을 개발하고 성능을 경쟁하는 대회
😎 2. 데이터 구성
- 학습 데이터셋 (3개)
- 파일명 : 'train1.csv', 'train2.csv', 'train3.csv'
- 설명 : 정상적인 운영 상황에서 수집된 데이터(각 파일별로 시간 연속성을 가짐)
- Column1 ('time') : 관측 시각
- Column2, 3, …, 80 ('C01', 'C02', …, 'C79') : 상태 관측 데이터
- 검증 데이터셋 (1개)
- 파일명 : 'validation.csv'
- 5가지 공격 상황에서 수집된 데이터(시간 연속성을 가짐)
- Column1 ('time') : 관측 시각
- Column2, 3, …, 80 ('C01', 'C02', …, 'C79') : 상태 관측 데이
- Column81 : 공격 라벨 (정상:'0', 공격:'1')
- 테스트 데이터셋 (4개)
- 파일명 : 'test1.csv', 'test2.csv', 'test3.csv', 'test4.csv'
- Column1 ('time') : 관측 시각
- Column2, 3, …, 80 ('C01', 'C02', …, 'C79') : 상태 관측 데이터
- HAI 2.0
- 파일명 : eTaPR-1.12-py3-none-any.whl
😎 3. 코드 구현
1️⃣ 데이터 로드
- !pip install /파일경로/eTaPR-1.12-py3-none-any.whl
!pip install /home/ubuntu/coding/220906/eTaPR-1.12-py3-none-any.whl
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import datetime
import time
from keras.models import Model, Sequential, load_model
import keras
from keras import optimizers
from keras.layers import Input,Bidirectional ,LSTM, Dense, Activation
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from sklearn.metrics import mean_squared_error
import tensorflow as tf
from keras.callbacks import EarlyStopping, ModelCheckpoint
from pathlib import Path
from datetime import timedelta
import dateutil
from tqdm.notebook import trange
from TaPR_pkg import etapr
- Data set
TRAIN_DATASET = sorted([x for x in Path("/home/ubuntu/coding/220906/HAI 2.0/training").glob("*.csv")])
TRAIN_DATASET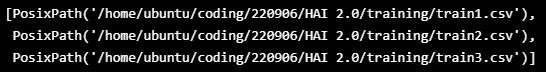
TEST_DATASET = sorted([x for x in Path("/home/ubuntu/coding/220906/HAI 2.0/testing").glob("*.csv")])
TEST_DATASET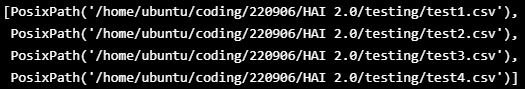
VALIDATION_DATASET = sorted([x for x in Path("/home/ubuntu/coding/220906/HAI 2.0/validation").glob("*.csv")])
VALIDATION_DATASET
def dataframe_from_csv(target):
return pd.read_csv(target).rename(columns=lambda x: x.strip())
def dataframe_from_csvs(targets):
return pd.concat([dataframe_from_csv(x) for x in targets])
- Data load
- Train dataset load
- 설명 : 정상적인 운영 상황에서 수집된 데이터(각 파일별로 시간 연속성을 가짐)
- Column1 ('time') : 관측 시각
- Column2, 3, …, 80 ('C01', 'C02', …, 'C79') : 상태 관측 데이터
- Train dataset load
TRAIN_DF_RAW = dataframe_from_csvs(TRAIN_DATASET)
TRAIN_DF_RAW
# 추후에 나는 데이터가 너무 커서 50000개로 줄였다.
# TRAIN_DF_RAW = dataframe_from_csvs(TRAIN_DATASET)
# TRAIN_DF_RAW = TRAIN_DF_RAW[40000:90000]
# TRAIN_DF_RAW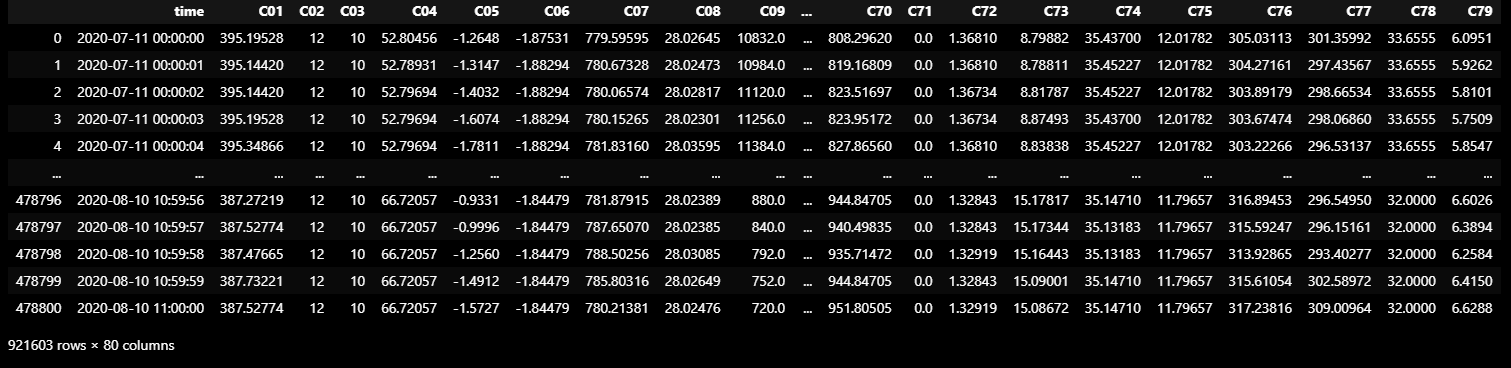
- 학습 데이터셋은 공격을 받지 않은 평상시 데이터이고 시간을 나타내는 필드인 time
- 나머지 필드는 모두 비식별화된 센서/액추에이터의 값
- 전체 데이터를 대상으로 이상을 탐지하므로 "attack" 필드만 사용
2️⃣ 데이터 전처리
- 학습 데이터셋에 있는 모든 센서/액추에이터 필드를 담고 있음
- 학습 데이터셋에 존재하지 않는 필드가 테스트 데이터셋에 존재하는 경우가 있음
- 학습 시 보지 못했던 필드에 대해서 테스트를 할 수 없으므로 학습 데이터셋을 기준으로 필드 이름
- 학습 데이터셋에서 최솟값 최댓값을 얻은 결과
TAG_MIN = TRAIN_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET].min()
TAG_MAX = TRAIN_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET].max()
- Min-Max Normalization (최소-최대 정규화)
- (X - MIN) / (MAX-MIN)
- 모든 feature에 대해 각각의 최소값 0, 최대값 1로, 그리고 다른 값들은 0과 1 사이의 값으로 변환
- 값이 전혀 변하지 않는 필드 경우 최솟값과 최댓값이 같을 것 -> 이런 필드를 모두 0
def normalize(df):
ndf = df.copy()
for c in df.columns:
if TAG_MIN[c] == TAG_MAX[c]:
ndf[c] = df[c] - TAG_MIN[c]
else:
ndf[c] = (df[c] - TAG_MIN[c]) / (TAG_MAX[c] - TAG_MIN[c])
return ndf
-
ewm (지수가중함수)
-
오래된 데이터에 지수감쇠를 적용하여 최근 데이터가 더 큰 영향을 끼지도록 가중치를 주는 함수
- 추가 메서드로 mean() 을 사용해서 지수가중평균으로 사용
-
사용법
-
df.ewm(com=None, span=None, halflife=None, alpha=None, min_periods=0, adjust=True, ignore_na=False, axis=0, times=None, method='single')
-
주로 가중치를 결정하는 요소는 alpha!! 평활계수(감쇠계수)====> com / span / halflife를 통해 자동 계산하도록 하거나, alpha를 통해 직접 설정
-
-
-
- exponential weighted function을 통과시킨 결과입니다. 센서에서 발생하는 noise를 smoothing 시켜주기
- Pandas Dataframe에 있는 값 중 1 초과의 값이 있는지, 0 미만의 값이 있는지, NaN이 있는지 점검
- np.any( ) : 배열의 데이터 중 조건과 맞는 데이터가 있으면 True, 전혀 없으면 False
def boundary_check(df):
x = np.array(df, dtype=np.float32)
return np.any(x > 1.0), np.any(x < 0), np.any(np.isnan(x))boundary_check(TRAIN_DF)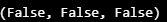
3️⃣ 학습 모델 설정 & 데이터 입출력 정의
- 베이스라인 모델은 Stacked RNN(GRU cells)을 이용해서 이상을 탐지
- 정상 데이터만 학습해야 하고, 정상 데이터에는 어떠한 label도 없으므로 unsupervised learning을 해야 함
- 슬라이딩 윈도우를 통해 시계열 데이터의 일부를 가져와서 해당 윈도우의 패턴을 기억
- 슬라이딩 윈도우는 90초(HAI는 1초마다 샘플링되어 있습니다)로 설정
- 모델의 입출력
- 입력 : 윈도우의 앞부분 89초에 해당하는 값
- 출력 : 윈도우의 가장 마지막 초(90번째 초)의 값
- 탐지 시에는 모델이 출력하는 값(예측값)과 실제로 들어온 값의 차를 보고 차이가 크면 이상으로 간주
- 많은 오차가 발생한다는 것은 기존에 학습 데이터셋에서 본 적이 없는 패턴이기 때문 (가정)
- HaiDataset : PyTorch의 Dataset 인터페이스를 정의한 것
- 데이터셋을 읽을 때는 슬라이딩 윈도우가 유효한 지 점검
- 정상적인 윈도우라면 원도우의 첫 시각과 마지막 시각의 차가 89초
- stride 파라미터 : 슬라이딩을 할 때 크기
- 전체 윈도우를 모두 학습할 수도 있지만, 시계열 데이터에서는 슬라이딩 윈도우를 1초씩 적용하면 이전 윈도우와 다음 윈도우의 값이 거의 같음
- 학습을 빠르게 마치기 위해 10초씩 건너뛰면서 데이터를 추출
- 슬라이딩 크기를 1로 설정하여 모든 데이터셋을 보게 하면 더 좋을 것
WINDOW_GIVEN = 59
WINDOW_SIZE= 60
def HaiDataset(timestamps, df, stride=1, attacks=None):
ts= np.array(timestamps)
tag_values=np.array(df,dtype=np.float32)
valid_idxs=[]
for L in trange(len(ts)-WINDOW_SIZE+1):
R = L + WINDOW_SIZE - 1
if dateutil.parser.parse(ts[R]) - dateutil.parser.parse(ts[L]) == timedelta(seconds=WINDOW_SIZE - 1):
valid_idxs.append(L)
valid_idxs=np.array(valid_idxs, dtype=np.int32)[::stride]
n_idxs = len(valid_idxs)
print("# of valid windows:", n_idxs)
if attacks is not None:
attacks = np.array(attacks, dtype=np.float32)
with_attack = True
else:
with_attack = False
timestamp, X, y, att = list(), list(), list(), list()
if with_attack:
for i in valid_idxs:
last=i+WINDOW_SIZE-1
seq_time, seq_x, seq_y, seq_attack = ts[last], tag_values[i:i+WINDOW_GIVEN], tag_values[last], attacks[last]
timestamp.append(seq_time)
X.append(seq_x)
y.append(seq_y)
att.append(seq_attack)
return np.array(timestamp), np.array(X), np.array(y), np.array(att)
else:
for i in valid_idxs:
last=i+WINDOW_SIZE-1
seq_time, seq_x, seq_y = ts[last], tag_values[i:i+WINDOW_GIVEN], tag_values[last]
timestamp.append(seq_time)
X.append(seq_x)
y.append(seq_y)
return np.array(timestamp), np.array(X), np.array(y)
- 훈련 데이터 나누기
ts, X_train, y_train = HaiDataset(TRAIN_DF_RAW[TIMESTAMP_FIELD], TRAIN_DF, stride=1)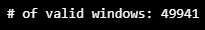
- 데이터셋이 잘 로드됨
- 모델은 3층 bidirectional GRU를 사용
- Hidden cell의 크기는 100
- Dropout은 사용하지 않음
HAI_DATASET_TRAIN = HaiDataset(TRAIN_DF_RAW[TIMESTAMP_FIELD], TRAIN_DF, stride=10)
HAI_DATASET_TRAIN[0]
- Validation dataset load
- Validation도 위와 같이 정규화
- 점검하기
VALIDATION_DF_RAW = dataframe_from_csvs(VALIDATION_DATASET)
VALIDATION_DF_RAW
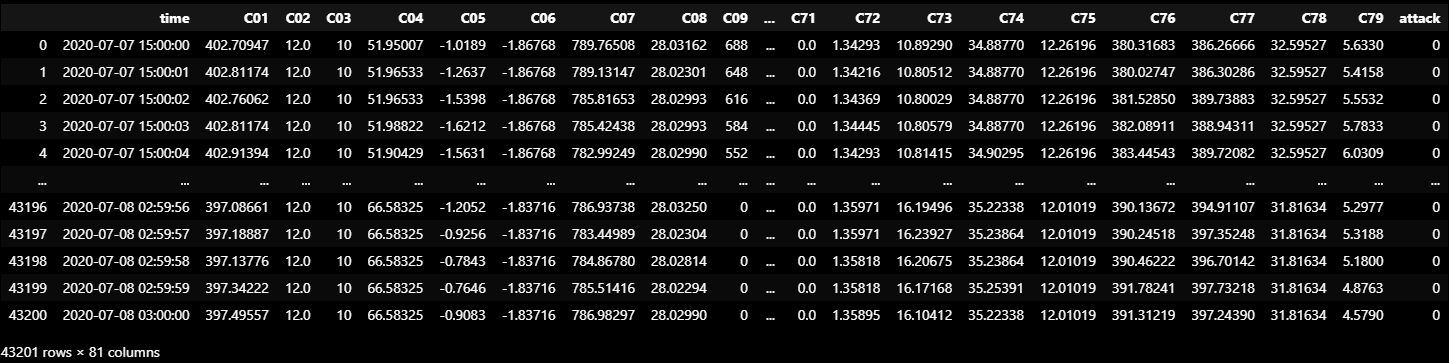
VALIDATION_DF = normalize(VALIDATION_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET])boundary_check(VALIDATION_DF)
ts, X_valid, y_valid, attack = HaiDataset(VALIDATION_DF_RAW[TIMESTAMP_FIELD], VALIDATION_DF, attacks=VALIDATION_DF_RAW[ATTACK_FIELD])
- EarlyStopping 지정
- 과적합을 방지하기 위해서는 EarlyStopping 이라는 콜백함수를 사용하여 적절한 시점에 학습을 조기 종료
더보기
EarlyStopping
EarlyStopping(monitor = 'val_loss', min_delta = 0, patience = 0, mode = 'auto')- monitor : 학습 조기종료를 위해 관찰
- val_loss 나 val_accuracy 가 주로 사용 (default : val_loss)
- min_delta : 개선되고 있다고 판단하기 위한 최소 변화량
- 변화량이 min_delta 보다 적은 경우에는 개선이 없다고 판단 (default = 0)
- patience : 개선이 안된다고 바로 종료시키지 않고, 개선을 위해 몇번의 에포크를 기다릴지 설정 (default = 0)
- mode : 관찰항목에 대해 개선이 없다고 판단하기 위한 기준을 설정
- monitor에서 설정한 항목이 val_loss 이면 값이 감소되지 않을 때 종료하여야 하므로 min 을 설정
- val_accuracy 의 경우에는 max를 설정 (default = auto)
- auto : monitor에 설정된 이름에 따라 자동으로 지정
- min : 관찰값이 감소하는 것을 멈출 때, 학습을 종료
- max: 관찰값이 증가하는 것을 멈출 때, 학습을 종료
- ModelCheckpoint 지정
더보기
ModelCheckpoint
tf.keras.callbacks.ModelCheckpoint(
filepath, monitor='val_loss', verbose=0, save_best_only=False,
save_weights_only=False, mode='auto', save_freq='epoch', options=None, **kwargs
)- 모델이 학습하면서 정의한 조건을 만족했을 때 Model의 weight 값을 중간 저장
- 학습시간이 꽤 오래걸린다면, 모델이 개선된 validation score를 도출해낼 때마다 weight를 중간 저장함
- 중간에 memory overflow나 crash가 나더라도 다시 weight를 불러와서 학습 이어나갈 수 있음 ( 시간 save)
| 인자 | 설명 |
| filepath | 모델을 저장할 경로를 입력 |
| monitor | 모델을 저장할 때, 기준이 되는 값을 지정 |
| verbose | 0, 1 1일 경우 모델이 저장 될 때, '저장되었습니다' 라고 화면에 표시 0일 경우 화면에 표시되는 것 없이 그냥 바로 모델이 저장 |
| save_best_only | True, False True 인 경우, monitor 되고 있는 값을 기준으로 가장 좋은 값으로 모델이 저장 False인 경우, 매 에폭마다 모델이 filepath{epoch}으로 저장 (model0, model1, model2....) |
| save_weights_only | True, False True인 경우, 모델의 weights만 저장 False인 경우, 모델 레이어 및 weights 모두 저장 |
| mode | 'auto', 'min', 'max' val_acc 인 경우, 정확도이기 때문에 클수록 좋음 (max를 입력) val_loss 인 경우, loss 값이기 때문에 값이 작을수록 좋음 (min을 입력) auto로 할 경우, 모델이 알아서 min, max를 판단하여 모델을 저장 |
| save_freq | 'epoch' 또는 integer(정수형 숫자) 'epoch'을 사용할 경우, 매 에폭마다 모델이 저장 integer을 사용할 경우, 숫자만큼의 배치를 진행되면 모델이 저장 |
| options | tf.train.CheckpointOptions를 옵션으로 주기 분산환경에서 다른 디렉토리에 모델을 저장하고 싶을 경우 사용 |
es = EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=4)
mc = ModelCheckpoint('best_LSTM_model_60seq_TEST.h5', monitor='val_loss', mode='min', verbose=1, save_best_only=True)
4️⃣ Training Model
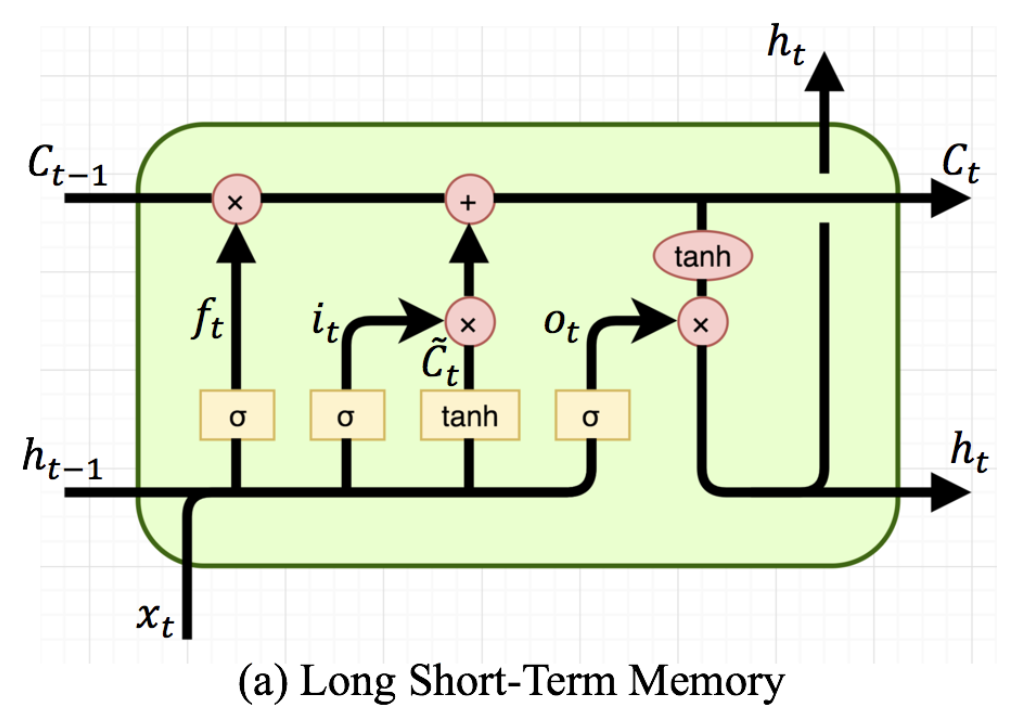
- LSTM (Long Short-Term Memory)
- 단기 메모리와 장기 메모리를 나눠 학습 후, 두 메모리를 병합해 이벤트 확률을 예측 → 과거의 정보를 훨씬 잘 반영
- LSTM은 게이트라는 개념으로 선별 기억을 확보
- C(Cell state)는 장기 기억 상태
- H(Hidden State)는 단기 기억 상태
- x나 h, C는 모두 데이터의 벡터
## LSTM Model define
n_features = TRAIN_DF.shape[1]
inputs=Input(shape=(WINDOW_GIVEN , n_features))
first=Bidirectional(LSTM(100, return_sequences=True))(inputs)
second=Bidirectional(LSTM(100, return_sequences=True))(first)
thrid=Bidirectional(LSTM(100))(second)
lstm_out=Dense(n_features)(thrid)
aux_input = Input(shape=(n_features,), name='aux_input')
outputs = keras.layers.add([lstm_out, aux_input])
model_60seq=Model(inputs=[inputs, aux_input], outputs=outputs)
model_60seq.compile(loss='mean_squared_error', optimizer='Adam')
model_60seq.summary()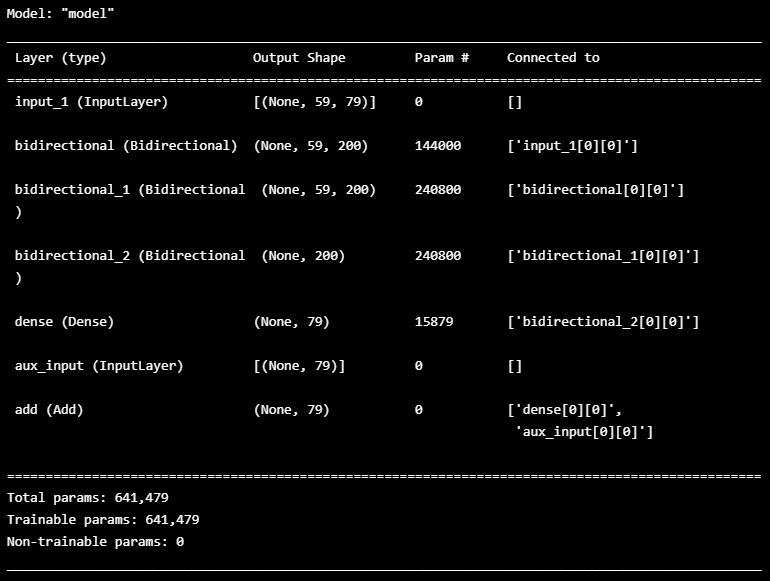
- aux_train으로 리스트 정리
- train, valid 둘다 ㄱ ㄱ
aux_train=[]
for i in range(len(X_train)):
aux_train.append(X_train[i][0])
aux_train=np.array(aux_train)aux_valid=[]
for i in range(len(X_valid)):
aux_valid.append(X_valid[i][0])
aux_valid=np.array(aux_valid)
- 모델 훈련
- earlystoppoing!
hist=model_60seq.fit([X_train, aux_train], y_train, batch_size=512, epochs=32, shuffle=True,
callbacks=[es, mc], validation_data=([X_valid,aux_valid],y_valid))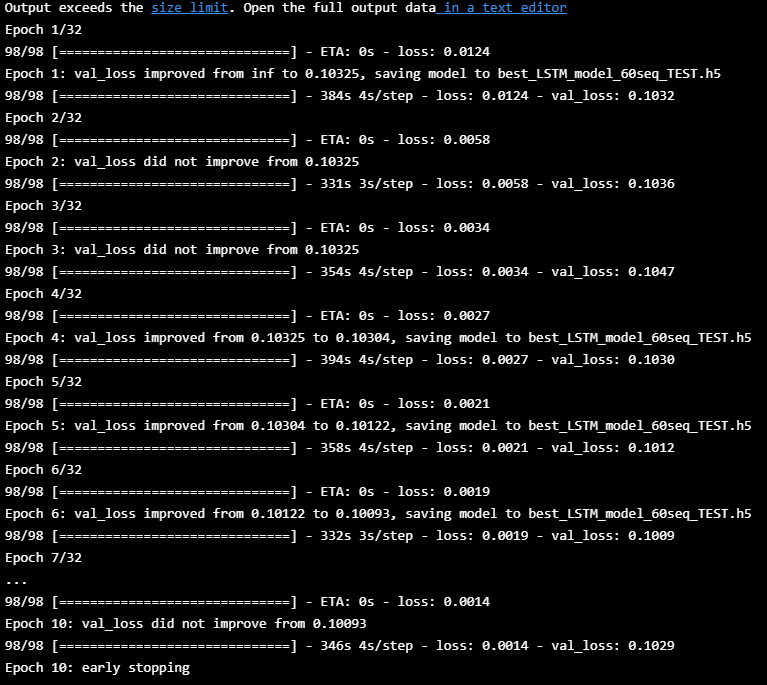
- 모델저장
model_60seq.save('best_LSTM_model_60seq.h5') #keras h5
5️⃣ Skip learning
## model load
model_60seq = load_model('best_LSTM_model_60seq.h5')- Validation 예측
## Validation set Prediction
y_pred=model_60seq.predict([X_valid,aux_valid])tmp=[]
for i in range(len(y_pred)):
tmp.append(abs(y_valid[i]-y_pred[i]))
- anomaly score 구하기
ANOMALY_SCORE=np.mean(tmp,axis=1)- threshold 이용해 label 값 넣기
def put_labels(distance, threshold):
xs = np.zeros_like(distance)
xs[distance > threshold] = 1
return xs- labels
THRESHOLD = 0.027
LABELS = put_labels(ANOMALY_SCORE, THRESHOLD)
LABELS, LABELS.shape
- attack labels
ATTACK_LABELS = put_labels(np.array(VALIDATION_DF_RAW[ATTACK_FIELD]), threshold=0.5)
ATTACK_LABELS, ATTACK_LABELS.shape
- what's this.?
def fill_blank(check_ts, labels, total_ts):
def ts_generator():
for t in total_ts:
yield dateutil.parser.parse(t)
def label_generator():
for t, label in zip(check_ts, labels):
yield dateutil.parser.parse(t), label
g_ts = ts_generator()
g_label = label_generator()
final_labels = []
try:
current = next(g_ts)
ts_label, label = next(g_label)
while True:
if current > ts_label:
ts_label, label = next(g_label)
continue
elif current < ts_label:
final_labels.append(0)
current = next(g_ts)
continue
final_labels.append(label)
current = next(g_ts)
ts_label, label = next(g_label)
except StopIteration:
return np.array(final_labels, dtype=np.int8)- final labels
%%time
FINAL_LABELS = fill_blank(ts, LABELS, np.array(VALIDATION_DF_RAW[TIMESTAMP_FIELD]))
FINAL_LABELS.shape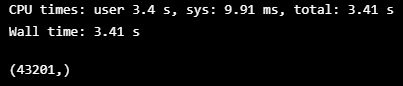
ATTACK_LABELS.shape[0] == FINAL_LABELS.shape[0]
# 성능 평가도구 eTaPR(enhanced Time-series aware Precision and Recall)
TaPR = etapr.evaluate(anomalies=ATTACK_LABELS, predictions=FINAL_LABELS)
print(f"F1: {TaPR['f1']:.3f} (TaP: {TaPR['TaP']:.3f}, TaR: {TaPR['TaR']:.3f})")
print(f"# of detected anomalies: {len(TaPR['Detected_Anomalies'])}")
print(f"Detected anomalies: {TaPR['Detected_Anomalies']}")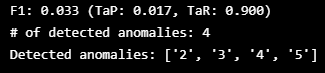
6️⃣ Moving Average
## MOVING AVERAGE
seq60_10mean=[]
for idx in range(len(ANOMALY_SCORE)):
if idx >= 10:
seq60_10mean.append((ANOMALY_SCORE[idx-10:idx].mean()+ANOMALY_SCORE[idx])/2)
else:
seq60_10mean.append(ANOMALY_SCORE[idx])
seq60_10mean=np.array(seq60_10mean)
print(seq60_10mean.shape)
THRESHOLD = 0.019
LABELS = put_labels(seq60_10mean, THRESHOLD)
LABELS, LABELS.shape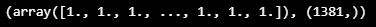
%%time
FINAL_LABELS = fill_blank(ts, LABELS, np.array(VALIDATION_DF_RAW[TIMESTAMP_FIELD]))
FINAL_LABELS.shape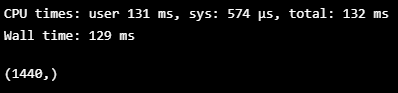
ATTACK_LABELS.shape[0] == FINAL_LABELS.shape[0]
TaPR = etapr.evaluate(anomalies=ATTACK_LABELS, predictions=FINAL_LABELS)
print(f"F1: {TaPR['f1']:.3f} (TaP: {TaPR['TaP']:.3f}, TaR: {TaPR['TaR']:.3f})")
print(f"# of detected anomalies: {len(TaPR['Detected_Anomalies'])}")
print(f"Detected anomalies: {TaPR['Detected_Anomalies']}")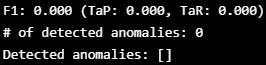
- test data 위와 같이
############### Test data Start
TEST_DF_RAW = dataframe_from_csvs(TEST_DATASET)
TEST_DF_RAW = TEST_DF_RAW[:11960]
TEST_DF_RAW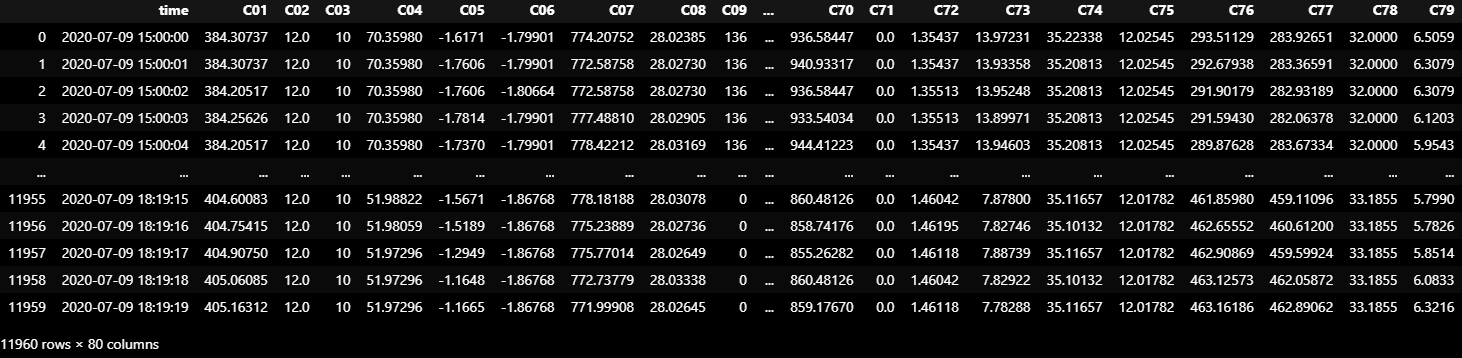
TEST_DF = normalize(TEST_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET]).ewm(alpha=0.9).mean()
TEST_DF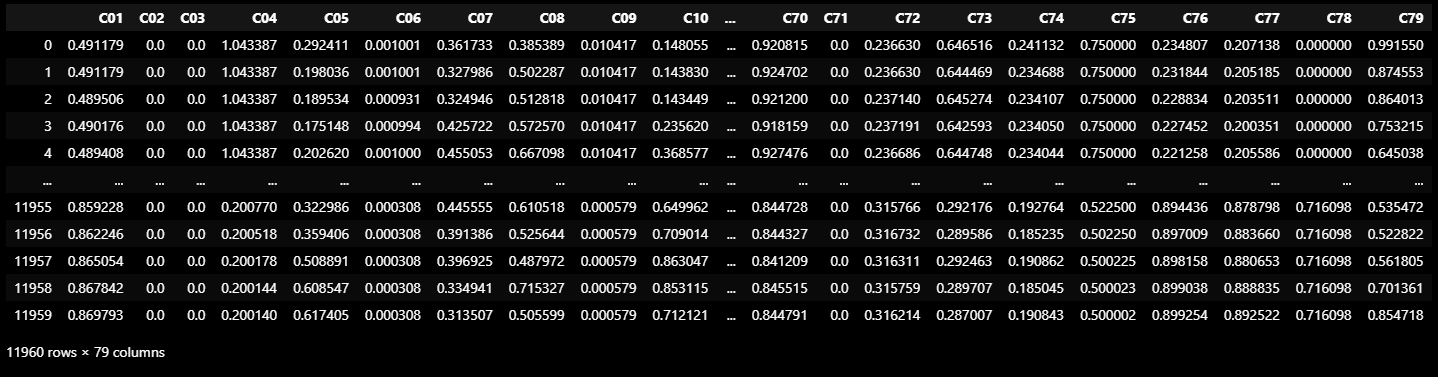
boundary_check(TEST_DF)
CHECK_TS, X_test, y_test = HaiDataset(TEST_DF_RAW[TIMESTAMP_FIELD], TEST_DF, attacks=None)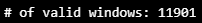
aux_test=[]
for i in range(len(X_test)):
aux_test.append(X_test[i][0])
aux_test=np.array(aux_test)## Model Prediction
y_pred=model_60seq.predict([X_test,aux_test])tmp=[]
for i in range(len(y_test)):
tmp.append(abs(y_test[i]-y_pred[i]))ANOMALY_SCORE=np.mean(tmp,axis=1)# Moving Average
seq60_10mean=[]
for idx in range(len(ANOMALY_SCORE)):
if idx >= 10:
seq60_10mean.append((ANOMALY_SCORE[idx-10:idx].mean()+ANOMALY_SCORE[idx])/2)
else:
seq60_10mean.append(ANOMALY_SCORE[idx])
seq60_10mean=np.array(seq60_10mean)
print(seq60_10mean.shape)
### Threshold Setting
THRESHOLD=0.019
LABELS_60seq = put_labels(seq60_10mean, THRESHOLD)
LABELS_60seq, LABELS_60seq.shape
submission = pd.read_csv('/home/ubuntu/coding/220906/HAI 2.0/sample_submission.csv')
submission.index = submission['time']
submission.loc[CHECK_TS,'attack'] = LABELS_60seq
submission
submission.to_csv('SILab_Moving_Average.csv', index=False)
7️⃣ Gray Aare Smoothing
submission = pd.read_csv('/home/ubuntu/coding/220906/HAI 2.0/sample_submission.csv')
submission.index = submission['time']
submission['attack_1']=0
submission['anomaly_score']=0
submission.loc[CHECK_TS,'anomaly_score'] = seq60_10mean
submission.loc[CHECK_TS,'attack_1'] = LABELS_60seq
LABELS_60seq=submission['attack_1']
seq60_10mean=submission['anomaly_score']def Gray_Area(attacks):
start = [] # start point
finish = [] # finish point
c = [] # count
com = 0
count = 0
for i in range(1, len(attacks)):
if attacks[i - 1] != attacks[i]:
if com == 0:
start.append(i)
count = count + 1
com = 1
elif com == 1:
finish.append(i - 1)
c.append(count)
count = 0
start.append(i)
count = count + 1
else:
count = count + 1
finish.append(len(attacks) - 1)
c.append(finish[len(finish) - 1] - start[len(start) - 1] + 1)
for i in range(0, len(start)):
if c[i] < 10:
s = start[i]
f = finish[i] + 1
g1 = [1 for i in range(c[i])] # Temp Attack list
g0 = [0 for i in range(c[i])] # Temp Normal List
if attacks[start[i] - 1] == 1:
attacks[s:f] = g1 # change to attack
else:
attacks[s:f] = g0 # change to normal
return attacksgray_LABELS_60seq=Gray_Area(LABELS_60seq)submission = pd.read_csv('/home/ubuntu/coding/220906/HAI 2.0/sample_submission.csv')
submission.index = submission['time']
submission['attack']=gray_LABELS_60seq
submission
submission.to_csv('SILab_Gray_Area_Smoothing.csv', index=False)
8️⃣ Attack Point and Finish Point Policy
WINDOW_GIVEN = 9
WINDOW_SIZE= 10ts, X_train, y_train = HaiDataset(TRAIN_DF_RAW[TIMESTAMP_FIELD], TRAIN_DF, stride=1)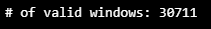
ts, X_valid, y_valid, attack = HaiDataset(VALIDATION_DF_RAW[TIMESTAMP_FIELD],
VALIDATION_DF, attacks=VALIDATION_DF_RAW[ATTACK_FIELD])
## LSTM Model define
n_features = TRAIN_DF.shape[1]
inputs=Input(shape=(WINDOW_GIVEN , n_features))
first=Bidirectional(LSTM(100, return_sequences=True))(inputs)
second=Bidirectional(LSTM(100, return_sequences=True))(first)
thrid=Bidirectional(LSTM(100))(second)
lstm_out=Dense(n_features)(thrid)
aux_input = Input(shape=(n_features,), name='aux_input')
outputs = keras.layers.add([lstm_out, aux_input])
model_10seq=Model(inputs=[inputs, aux_input], outputs=outputs)
model_10seq.compile(loss='mean_squared_error', optimizer='Adam')
model_10seq.summary()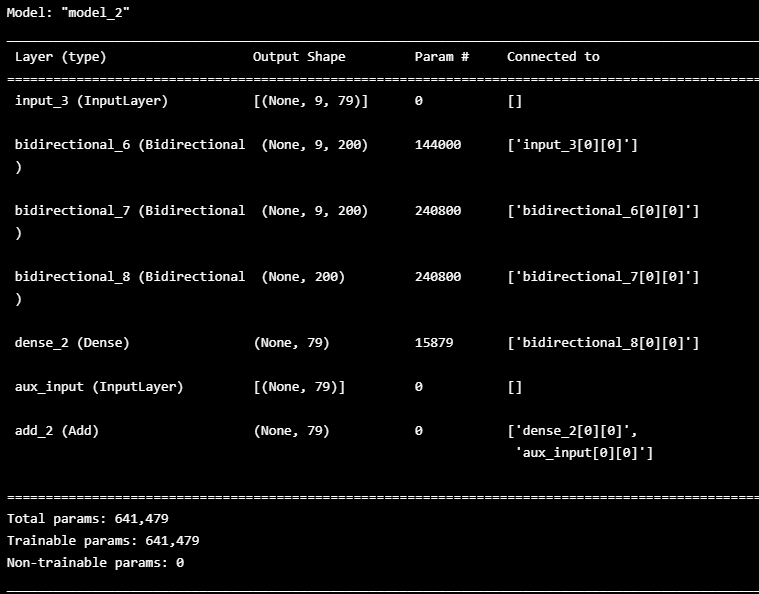
aux_train=[]
for i in range(len(X_train)):
aux_train.append(X_train[i][0])
aux_train=np.array(aux_train)aux_valid=[]
for i in range(len(X_valid)):
aux_valid.append(X_valid[i][0])
aux_valid=np.array(aux_valid)hist=model_10seq.fit([X_train, aux_train], y_train, batch_size=512, epochs=32, shuffle=True,
callbacks=[es, mc], validation_data=([X_valid,aux_valid],y_valid))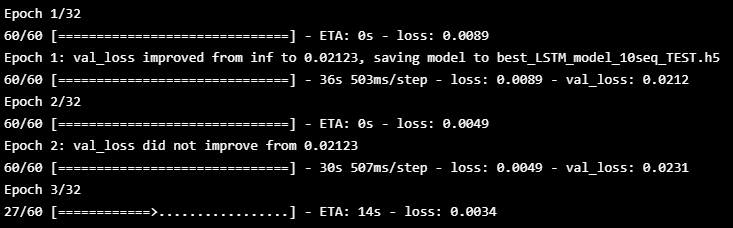
model_10seq.save('best_LSTM_model_10seq.h5') #keras h5## model load
model_10seq = load_model('best_LSTM_model_10seq.h5')CHECK_TS, X_test, y_test = HaiDataset(TEST_DF_RAW[TIMESTAMP_FIELD], TEST_DF, attacks=None)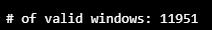
aux_test=[]
for i in range(len(X_test)):
aux_test.append(X_test[i][0])
aux_test=np.array(aux_test)## Model Prediction
y_pred=model_10seq.predict([X_test,aux_test])tmp=[]
for i in range(len(y_test)):
tmp.append(abs(y_test[i]-y_pred[i]))ANOMALY_SCORE=np.mean(tmp,axis=1)# Moving Average
seq10_10mean=[]
for idx in range(len(ANOMALY_SCORE)):
if idx >= 10:
seq10_10mean.append((ANOMALY_SCORE[idx-10:idx].mean()+ANOMALY_SCORE[idx])/2)
else:
seq10_10mean.append(ANOMALY_SCORE[idx])
seq10_10mean=np.array(seq10_10mean)
print(seq10_10mean.shape)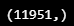
### Threshold Setting
THRESHOLD=0.008
LABELS_10seq = put_labels(seq10_10mean, THRESHOLD)
LABELS_10seq, LABELS_10seq.shape
submission = pd.read_csv('./HAI 2.0/sample_submission.csv')
submission.index = submission['time']
submission['attack_1']=0
submission.loc[CHECK_TS,'attack_1'] = LABELS_10seq
LABELS_10seq=submission['attack_1']gray_LABELS_10seq=Gray_Area(LABELS_10seq)Queue=[0 for i in range(59)]
Label=[]
for i in range(len(gray_LABELS_60seq)):
Queue.append(gray_LABELS_60seq[i])
N=Queue.count(1)
if N>=60:
if seq60_10mean[i-100:i].max() > 0.1:
Label.append(gray_LABELS_10seq[i])
else:
Label.append(gray_LABELS_60seq[i])
elif N>=1:
Label.append(gray_LABELS_60seq[i])
else:
Label.append(gray_LABELS_60seq[i])
del Queue[0]
Label=np.array(Label)submission = pd.read_csv('/home/ubuntu/coding/220906/HAI 2.0/sample_submission.csv')
submission.index = submission['time']
submission['attack']=Label
submission
728x90
반응형
'👩💻 인공지능 (ML & DL) > Serial Data' 카테고리의 다른 글
| 시계열 모델 ARIMA 2 (자기회귀 집적 이동 평균) (0) | 2022.09.08 |
|---|---|
| [DACON] HAICon2020 산업제어시스템 보안위협 탐지 AI & 비지도 기반 Autoencoder (0) | 2022.09.08 |
| [이상 탐지] ML for Time Series & windows (0) | 2022.09.07 |
| ADTK (Anomaly detection toolkit) 시계열 이상탐지 오픈소스 (0) | 2022.09.06 |
| TimeSeries DeepLearing [Encoder-Decoder LSTM (=seq2seq)] (0) | 2022.09.05 |
Comments




