😎 공부하는 징징알파카는 처음이지?
[DACON] HAICon2020 산업제어시스템 보안위협 탐지 AI & 비지도 기반 Autoencoder 본문
👩💻 인공지능 (ML & DL)/Serial Data
[DACON] HAICon2020 산업제어시스템 보안위협 탐지 AI & 비지도 기반 Autoencoder
징징알파카 2022. 9. 8. 13:14728x90
반응형
220908 작성
<본 블로그는 dacon 대회에서의 데이크루 2기 Team Zoo 팀 코드와 dacon의 HAI 2.0 Baseline 글을 참고해서 공부하며 작성하였습니다 :-) >
https://dacon.io/codeshare/5141?dtype=recent
[Team Zoo] 특별편 4. 비지도학습 기반의 이상탐지 활용(feat. 시계열)
dacon.io
https://dacon.io/competitions/official/235624/codeshare/1570?page=1&dtype=recent
HAI 2.0 Baseline
HAICon2020 산업제어시스템 보안위협 탐지 AI 경진대회
dacon.io
1️⃣ Libraries & Data Load
!pip install /파일경로/eTaPR-1.12-py3-none-any.whl
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.preprocessing import StandardScaler
from sklearn import metrics
from tqdm.notebook import trange
from TaPR_pkg import etapr
from pathlib import Path
import time
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import load_model
from tensorflow.keras.callbacks import EarlyStopping
2️⃣ Data Preprocessing
1) Data load
TRAIN_DATASET = sorted([x for x in Path("/home/ubuntu/coding/220906/HAI 2.0/training").glob("*.csv")])
TRAIN_DATASET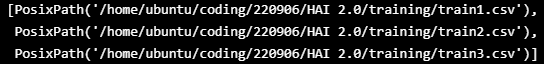
TEST_DATASET = sorted([x for x in Path("/home/ubuntu/coding/220906/HAI 2.0/testing").glob("*.csv")])
TEST_DATASET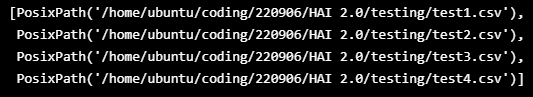
VALIDATION_DATASET = sorted([x for x in Path("/home/ubuntu/coding/220906/HAI 2.0/validation").glob("*.csv")])
VALIDATION_DATASET
def dataframe_from_csv(target):
return pd.read_csv(target, engine='python').rename(columns=lambda x: x.strip())
def dataframe_from_csvs(targets):
return pd.concat([dataframe_from_csv(x) for x in targets])TRAIN_DF_RAW = dataframe_from_csvs(TRAIN_DATASET)
TRAIN_DF_RAW = TRAIN_DF_RAW[:30720]
TRAIN_DF_RAW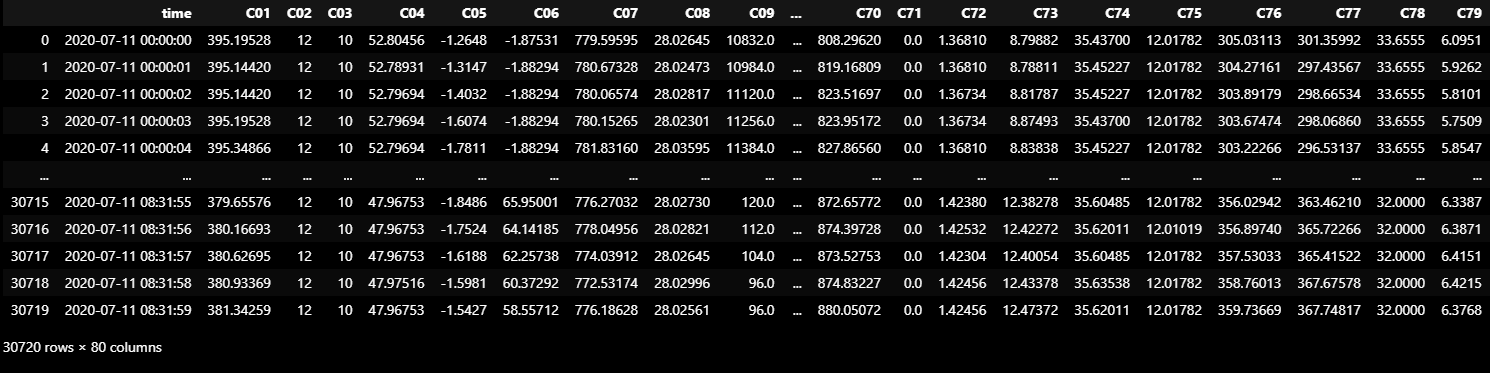
2) Variables setting
- 전체 데이터를 대상으로 이상을 탐지하므로 "attack" 필드만 사용
- VALID_COLUMNS_IN_TRAIN_DATASET은 학습 데이터셋에 있는 모든 센서/액추에이터 필드
- 학습 시 보지 못했던 필드에 대해서 테스트를 할 수 없으므로 학습 데이터셋을 기준으로 필드 이름
TIMESTAMP_FIELD = "time"
IDSTAMP_FIELD = 'id'
ATTACK_FIELD = "attack"
VALID_COLUMNS_IN_TRAIN_DATASET = TRAIN_DF_RAW.columns.drop([TIMESTAMP_FIELD])
VALID_COLUMNS_IN_TRAIN_DATASET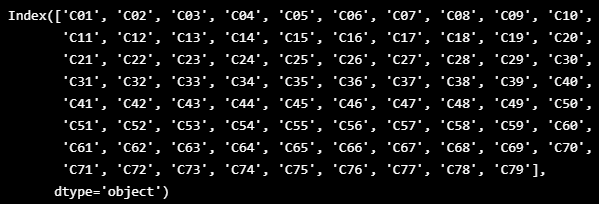
3) Data Normalization
- Min-Max Normalization (최소-최대 정규화)
- (X - MIN) / (MAX-MIN)
- 모든 feature에 대해 각각의 최소값 0, 최대값 1로, 그리고 다른 값들은 0과 1 사이의 값으로 변환
TAG_MIN = TRAIN_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET].min()
TAG_MAX = TRAIN_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET].max()def normalize(df):
ndf = df.copy()
for c in df.columns:
if TAG_MIN[c] == TAG_MAX[c]:
ndf[c] = df[c] - TAG_MIN[c]
else:
ndf[c] = (df[c] - TAG_MIN[c]) / (TAG_MAX[c] - TAG_MIN[c])
return ndfTRAIN_DF = normalize(TRAIN_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET])
- Pandas Dataframe에 있는 값 중 1 초과의 값이 있는지, 0 미만의 값이 있는지, NaN이 있는지 점검
- np.any( ) : 배열의 데이터 중 조건과 맞는 데이터가 있으면 True, 전혀 없으면 False
def boundary_check(df):
x = np.array(df, dtype=np.float32)
print(x)
return np.any(x > 1.0), np.any(x < 0), np.any(np.isnan(x))boundary_check(TRAIN_DF)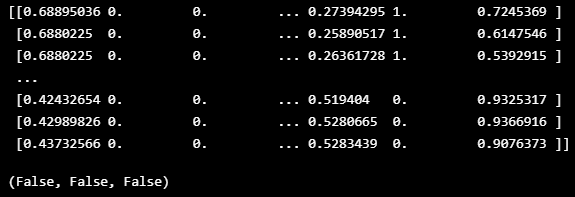
3️⃣ Model
- Autoencoder
- 주로 이미지의 생성이나 복원에 많이 사용
- 정상적인 이미지로 모델 학습 후 비정상적인 이미지를 넣어 이를 디코딩 하게 되면 정상 이미지 특성과 디코딩 된 이미지 간의 차이인 재구성 손실(Reconstruction Error)를 계산
- 재구성 손실이 낮은 부분은 정상(normal), 재구성 손실이 높은 부분은 이상(Abnormal)로 판단
- Autoencoder의 레이어를 LSTM으로 구성하여 시퀸스 학습이 가능
- !D-Convolution layer를 적용하여 timestamp와 feature 정보를 세밀하게 이동하면서 학습
- Encoder-Decoder LSTM (=seq2seq)
- input도 sequencial 데이터, output도 sequencial 데이터
- (문제) input과 output의 sequence 길이가 다를 수 있음
- (해결) Encoding : 여러 길이의 input을 고정 길이 벡터로 변환
- Encoder-Decoder LSTM 모델은 다양한 길이의 시계열 입력 데이터를 받아, 다양한 길이의 시계열 출력 데이터를 만들 수 있음
- LSTM Autoencoder는 다양한 길이의 시계열 input 데이터를 고정 길이 벡터로 압축해 Decoder에 입력으로 전달해줌
- 입력 데이터를 encoded feature vector로 변환하는 과정이 있음
def temporalize(X, y, timesteps):
output_X = []
output_y = []
for i in range(len(X) - timesteps - 1):
t = []
for j in range(1, timesteps + 1):
t.append(X[[(i + j + 1)], :])
output_X.append(t)
output_y.append(y[i + timesteps + 1])
return np.squeeze(np.array(output_X)), np.array(output_y)train = np.array(TRAIN_DF)
x_train = train.reshape(train.shape[0], 1, train.shape[1])
x_train.shape
▶ Model details
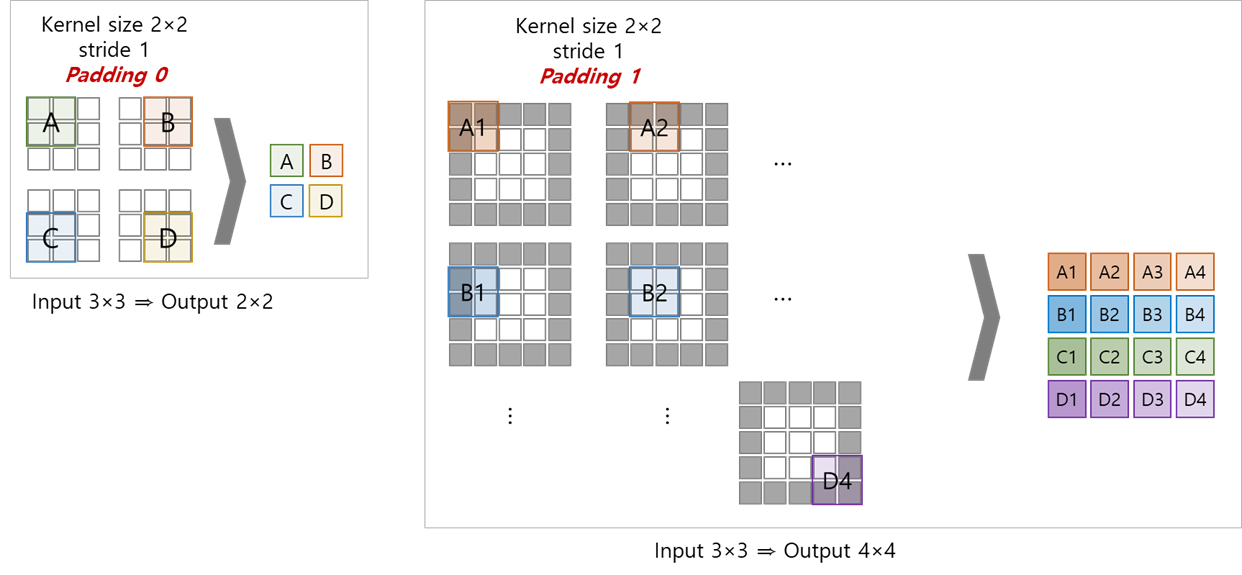
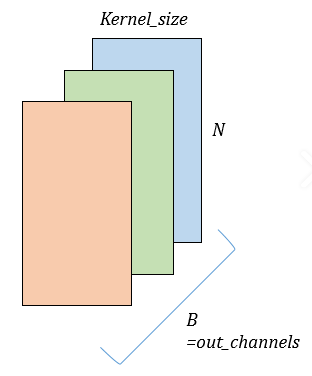
- Conv1D
- filters : 컨볼루션 연산의 output 출력 수
- kernel_size : timestamp를 얼마만큼 볼 것인가( = window_size)
- padding : 한 쪽 방향으로 얼마만큼 padding할 것인가
- dilation : kernel 내부에서 얼마만큼의 간격으로 kernel을 적용할 것인가
- stride : default = 1, 컨볼루션 레이어의 이동크기
- LSTM
- unit : 출력 차원층만 설정
- 모델의 구조
- Conv1D - Dense층 - LSTM - Dense층으로 encoder 와 decoder가 대칭이 되도록 설계
- 파라미터는 주로 filters, kernel_size, Dense, LSTM의 units 값을 조절
- Conv1D 레이어를 추가하거나 maxpooling과 같이 기존의 CNN 모델과 동일한 방식 적용 가능
def conv_auto_model(x):
n_steps = x.shape[1]
n_features = x.shape[2]
keras.backend.clear_session()
model = keras.Sequential(
[
layers.Input(shape=(n_steps, n_features)),
layers.Conv1D(filters=512, kernel_size=64, padding='same', data_format='channels_last',
dilation_rate=1, activation="linear"),
layers.Dense(128),
layers.LSTM(
units=64, activation="relu", name="lstm_1", return_sequences=False
),
layers.Dense(64),
layers.RepeatVector(n_steps),
layers.Dense(64),
layers.LSTM(
units=64, activation="relu", name="lstm_2", return_sequences=True
),
layers.Dense(128),
layers.Conv1D(filters=512, kernel_size=64, padding='same', data_format='channels_last',
dilation_rate=1, activation="linear"),
layers.TimeDistributed(layers.Dense(x.shape[2], activation='linear'))
]
)
return modelmodel1 = conv_auto_model(x_train)
model1.compile(optimizer='adam', loss='mse')
model1.summary()
4️⃣ Model fit
- epoch을 3으로 하고, earlystopping을 사용
early_stopping = EarlyStopping(monitor='val_loss', patience=5)
epochs = 3
batch = 64
# fit
history = model1.fit(x_train, x_train,
epochs=epochs, batch_size=batch,
validation_split=0.2, callbacks=[early_stopping]).history
model1.save('model1.h5')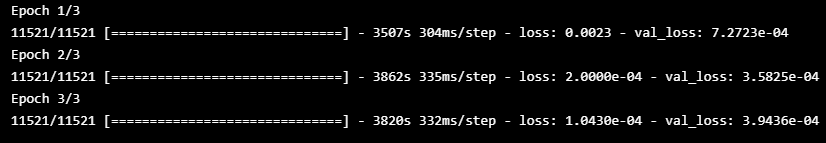
plt.plot(history['loss'], label='train loss')
plt.plot(history['val_loss'], label='valid loss')
plt.legend()
plt.xlabel('Epoch'); plt.ylabel('loss')
plt.show()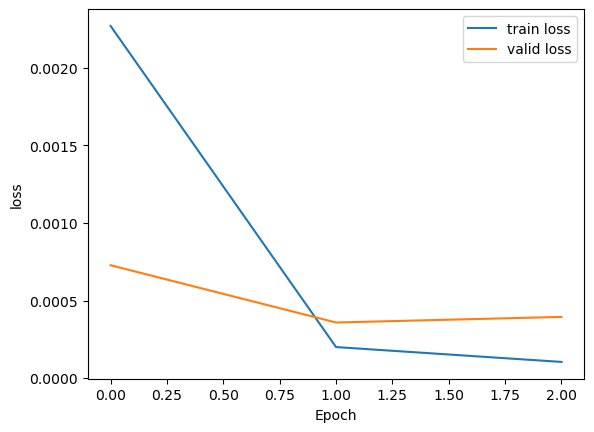
- model load
model1.save('best_AutoEncoder_model1.h5') #keras h5model = load_model('best_AutoEncoder_model1.h5')
5️⃣ Anomaly Detection
- 학습된 모델을 검증 데이터셋에 적용
VALIDATION_DF_RAW = dataframe_from_csvs(VALIDATION_DATASET)
VALIDATION_DF_RAW.to_csv('VALIDATION_DF_RAW.csv')
VALIDATION_DF_RAW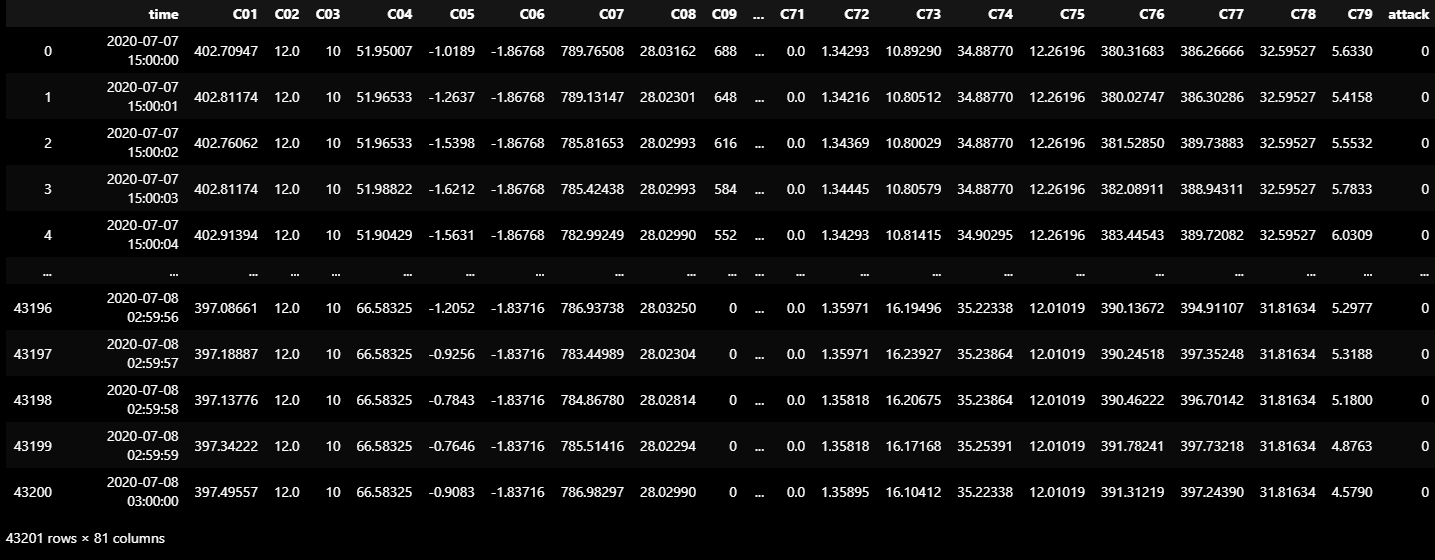
- 검증 데이터 셋에서는 정상 데이터를 기준으로 정규화를 진행
VALIDATION_DF = normalize(VALIDATION_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET])boundary_check(VALIDATION_DF)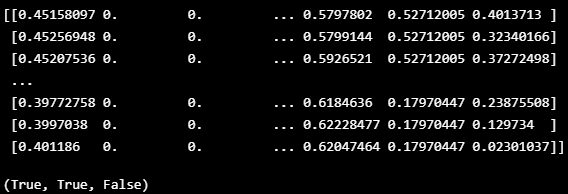
- 시각화로 일정 구간에서 0과 1 범위를 벗어나는 것을 확인
VALIDATION_DF.plot()
VALIDATION_DF['C75'].plot()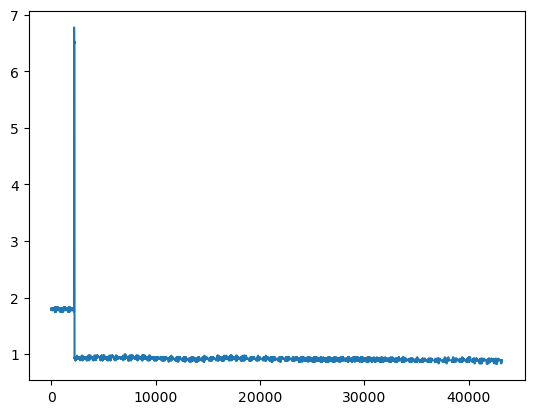
6️⃣ Data cleaning
- validation set에서 조금 더 정교하게 threshold 조절 및 결과를 확인하기 위해서 해당 변수의 값을 정상 범위에 맞게 임의로 조절
# valid 그래프를 보고 앞부분 정상인데 값이 튀는 변수가 있어서 조절
VALIDATION_DF['C75'][:2110] = 0.95val = np.array(VALIDATION_DF)
x_val = val.reshape(val.shape[0], 1, val.shape[1])
x_val.shape
- 모델의 결과가 3차원의 형태이기 때문에 복원된 결과와의 차이를 확인하기 위해서는 2차원으로 다시 바꾸기
def flatten(X):
flattened_X = np.empty((X.shape[0], X.shape[2])) # sample x features array.
for i in range(X.shape[0]):
flattened_X[i] = X[i, (X.shape[1]-1), :]
return(flattened_X)
def scale(X, scaler):
for i in range(X.shape[0]):
X[i, :, :] = scaler.transform(X[i, :, :])
return X
-
모델의 의해 재구성된 값을 실제 값과 차이를 구해서 재구성 손실(reconstruction error) 값을 구하기
-
정상인 경우 모델이 잘 학습되어 복원이 잘 되었기 때문에 reconstruction error 값이 작게 나올 것
-
공격인 경우 정규화된 값에서 0과 1을 벗어나기 때문에 reconstruction error 값이 크게 나올 것
-
start = time.time()
valid_x_predictions = model.predict(x_val)
print(valid_x_predictions.shape)
error = flatten(x_val) - flatten(valid_x_predictions)
print((flatten(x_val) - flatten(valid_x_predictions)).shape)
valid_mse = np.mean(np.power(flatten(x_val) - flatten(valid_x_predictions), 2), axis=1)
print(valid_mse.shape)
print(time.time()-start)
7️⃣ Precision Recall Curve
-
threshold의 경우 Recall과 Precision의 값이 교차되는 지점을 기준
error_df = pd.DataFrame({'Reconstruction_error': valid_mse, 'True_class':list(VALIDATION_DF_RAW['attack'])})
precision_rt, recall_rt, threshold_rt = metrics.precision_recall_curve(error_df['True_class'], error_df['Reconstruction_error'])
plt.figure(figsize=(8,5))
plt.plot(threshold_rt, precision_rt[1:], label='Precision')
plt.plot(threshold_rt, recall_rt[1:], label='Recall')
plt.xlabel('Threshold'); plt.ylabel('Precision/Recall')
plt.legend()
#plt.show()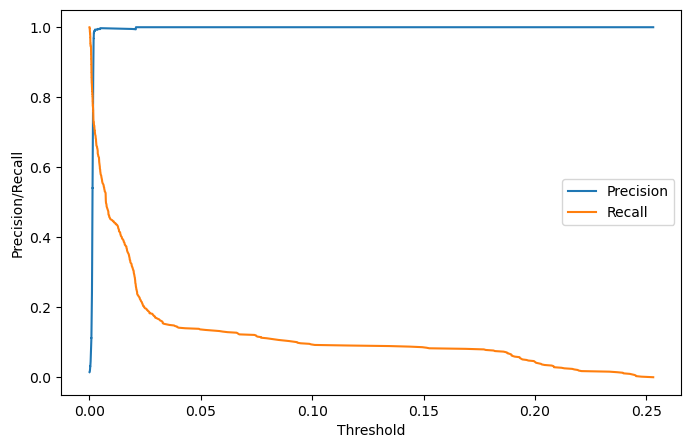
index_cnt = [cnt for cnt, (p, r) in enumerate(zip(precision_rt, recall_rt)) if p==r][0]
print('precision: ',precision_rt[index_cnt],', recall: ',recall_rt[index_cnt])
# fixed Threshold
threshold_fixed = threshold_rt[index_cnt]
print('threshold: ',threshold_fixed)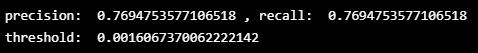
8️⃣ Predict Validation Data set
- reconstruction error 값이 작게 나왔고, 비정상인 구간은 확실하게 reconstruction error 값이 높게 나오는 것을 확인
error_df = pd.DataFrame({'Reconstruction_error': valid_mse ,
'True_class': list(VALIDATION_DF_RAW['attack'])})
groups = error_df.groupby('True_class')
fig, ax = plt.subplots(figsize=(20,20))
for name, group in groups:
ax.plot(group.index, group.Reconstruction_error, marker='o', ms=3.5, linestyle='',
label= "Break" if name == 1 else "Normal")
ax.hlines(threshold_fixed, ax.get_xlim()[0], ax.get_xlim()[1], colors="r", zorder=100, label='Threshold')
ax.legend()
plt.title("Reconstruction error for different classes")
plt.ylabel("Reconstruction error")
plt.xlabel("Data point index")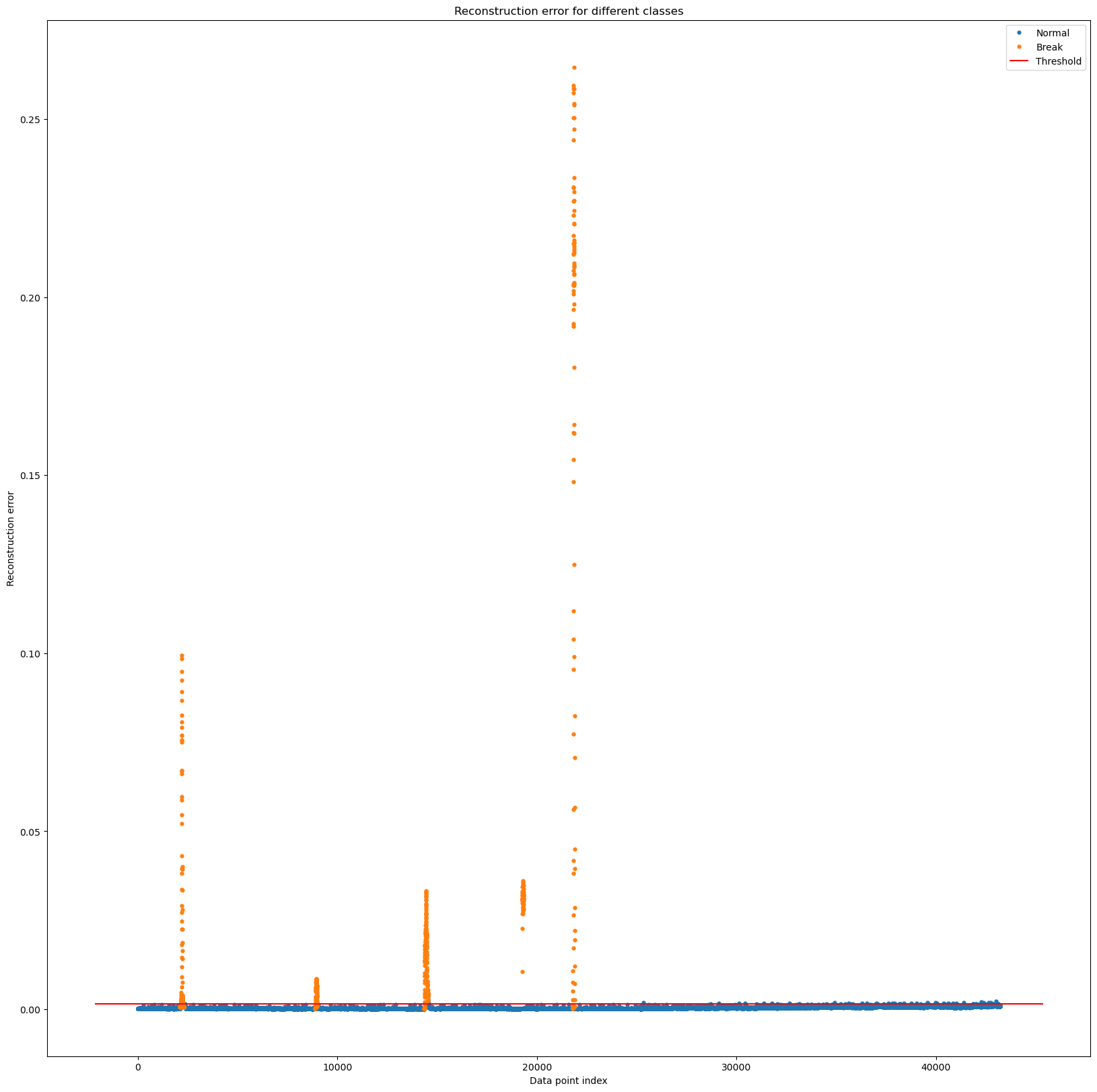
9️⃣ 이동평균
- 이동평균 값을 통해 정상인 구간은 평균적으로 더 낮게 하고, 비정상인 구간은 평균적으로 더 높은 값을 나타내도록 함
error_df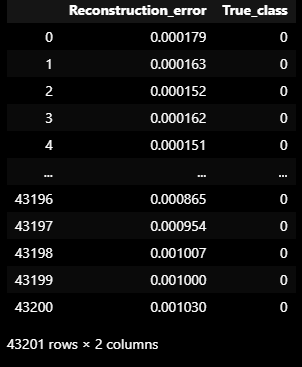
#이동평균
mean_window = error_df['Reconstruction_error'].rolling(50).mean()
window_error = mean_window.fillna(0)
window_error
- 확실하게 공격인 구간 잡기
window_error_df = pd.DataFrame({'Reconstruction_error': window_error ,
'True_class': list(VALIDATION_DF_RAW['attack'])})
groups = window_error_df.groupby('True_class')
fig, ax = plt.subplots(figsize=(20,20))
for name, group in groups:
ax.plot(group.index, group.Reconstruction_error, marker='o', ms=3.5, linestyle='',
label= "Break" if name == 1 else "Normal")
ax.hlines(threshold_fixed, ax.get_xlim()[0], ax.get_xlim()[1], colors="r", zorder=100, label='Threshold')
ax.legend()
plt.title("Reconstruction error for different classes")
plt.ylabel("Reconstruction error")
plt.xlabel("Data point index")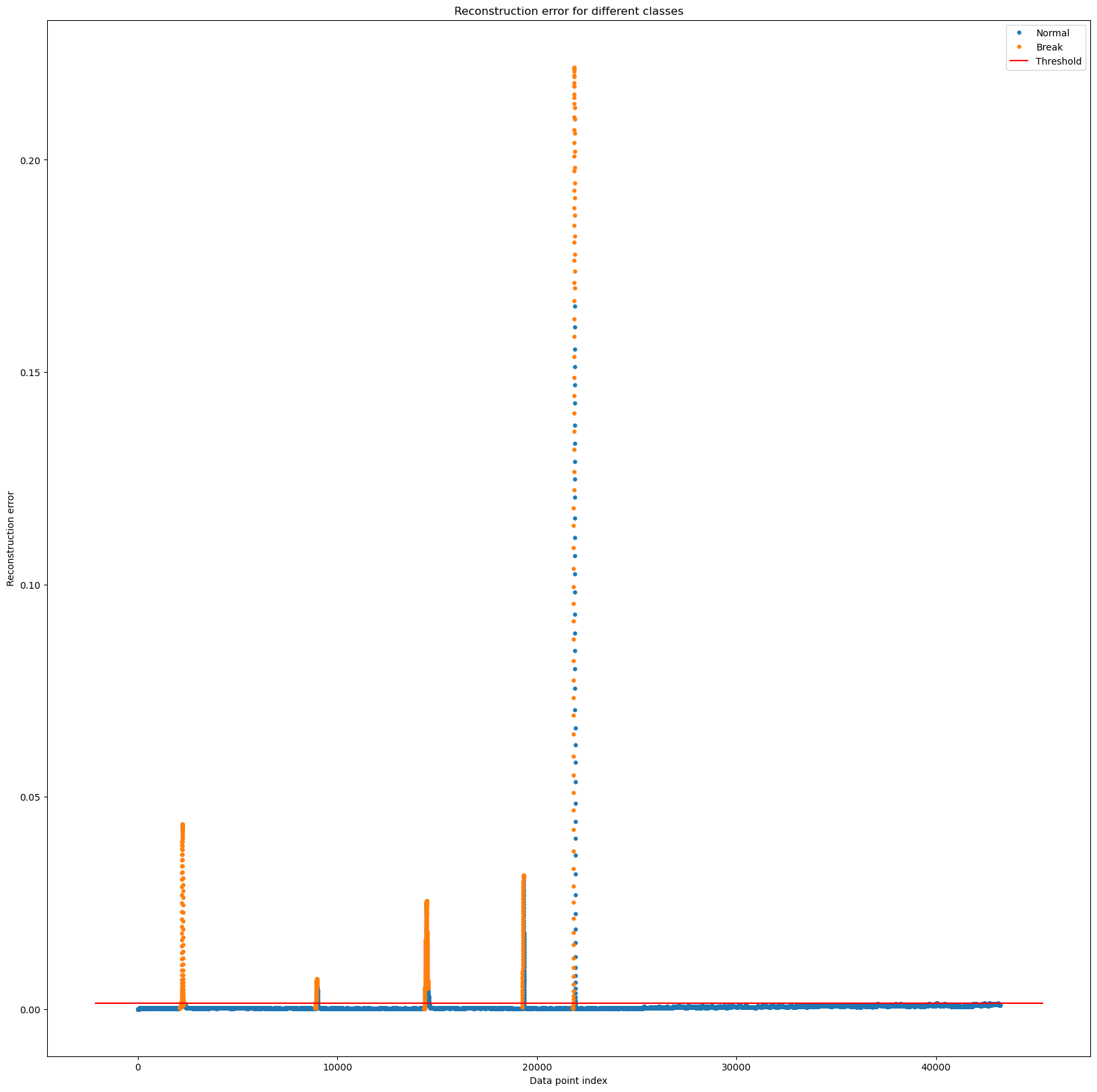
- threshold 값으로 validation set 의 결과를 확인
pred_y = [1 if e > threshold_fixed else 0 for e in window_error_df['Reconstruction_error'].values]
pred_y = np.array(pred_y)
pred_y.shape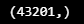
🔟 평가
ATTACK_LABELS = np.array(VALIDATION_DF_RAW[ATTACK_FIELD])
FINAL_LABELS = np.array(pred_y)
ATTACK_LABELS.shape[0] == FINAL_LABELS.shape[0]
TaPR = etapr.evaluate(anomalies=ATTACK_LABELS, predictions=FINAL_LABELS)
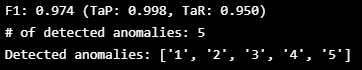
print(f"F1: {TaPR['f1']:.3f} (TaP: {TaPR['TaP']:.3f}, TaR: {TaPR['TaR']:.3f})")
print(f"# of detected anomalies: {len(TaPR['Detected_Anomalies'])}")
print(f"Detected anomalies: {TaPR['Detected_Anomalies']}")- 비정상 구간에서 특이했던 변수의 데이터 값을 조절하고 이동 평균 값을 활용하였을 때
- validation set에서 TaPR 점수가 99.8이라는 높은 점수가 나옴
1️⃣1️⃣ Predict Test Data set
- 위와 전처리, 정규화, 모델 동일
TEST_DF_RAW = dataframe_from_csvs(TEST_DATASET)
TEST_DF_RAW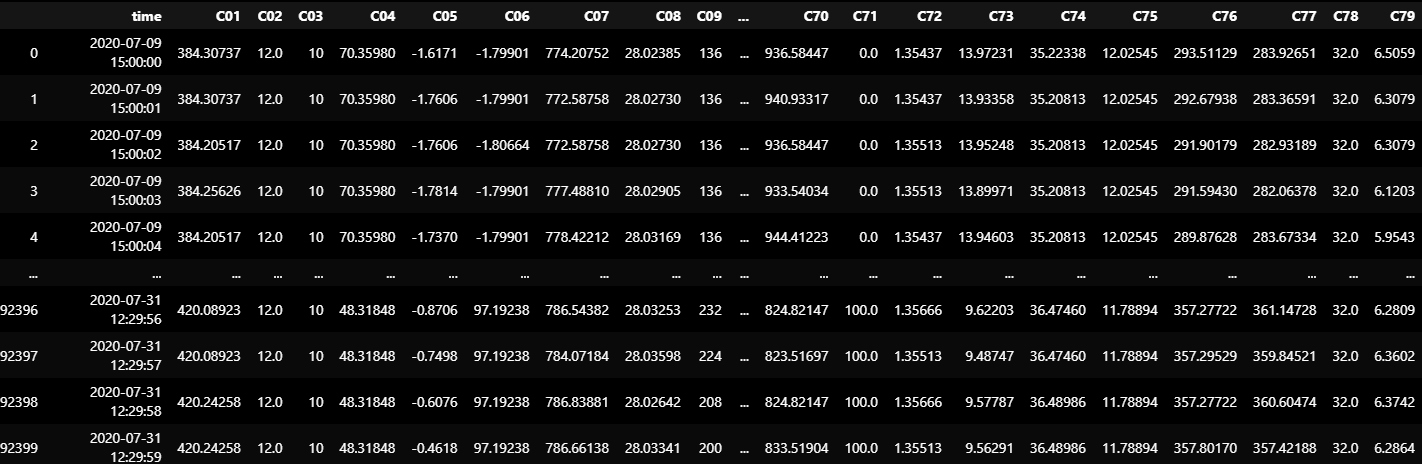
TEST_DF = normalize(TEST_DF_RAW[VALID_COLUMNS_IN_TRAIN_DATASET]).ewm(alpha=0.9).mean()
TEST_DF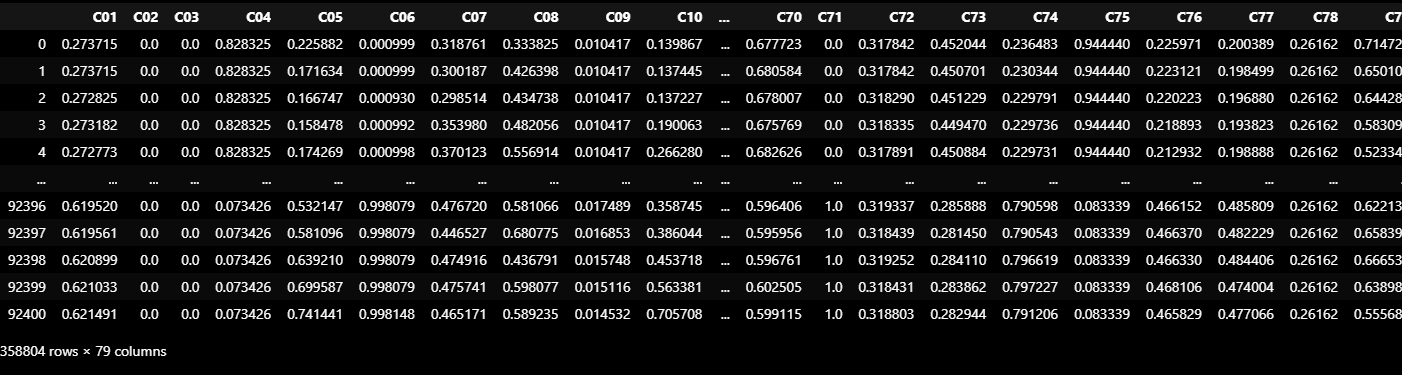
TEST_DF.plot()
boundary_check(TEST_DF)
test = np.array(TEST_DF)
x_test = test.reshape(test.shape[0], 1, test.shape[1])
x_test.shape
start = time.time()
test_x_predictions = model.predict(x_test)
print(test_x_predictions.shape)
test_mse = np.mean(np.power(flatten(x_test) - flatten(test_x_predictions), 2), axis=1)
print(test_mse.shape)
print(time.time()-start)
test_error = pd.DataFrame({'Reconstruction_error': test_mse})- 테스트 데이터 셋에서는 label 값을 알 수 없었기 때문에, 이동평균의 구간과 threshold 값을 조금씩 변경하면서 제출 후 결과를 보고 조절
movemean_test = pd.DataFrame({'Reconstruction_error': test_d})pred_y_test = [1 if e > 0.000425 else 0 for e in movemean_test['Reconstruction_error'].values]
pred_y_test = np.array(pred_y_test)
pred_y_test.shape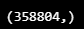
submission = pd.read_csv('HAI 2.0/sample_submission.csv')
submission.index = submission['time']
submission['attack'] = pred_y_testsubmission['attack'].value_counts()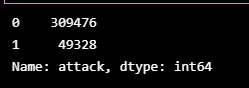
test_error_df = pd.DataFrame({'Reconstruction_error': test_d,
'True_class': list(submission['attack'])})
groups = test_error_df.groupby('True_class')
fig, ax = plt.subplots(figsize=(20,20))
for name, group in groups:
ax.plot(group.index, group.Reconstruction_error, marker='o', ms=3.5, linestyle='',
label= "Break" if name == 1 else "Normal")
ax.hlines(0.000425, ax.get_xlim()[0], ax.get_xlim()[1], colors="r", zorder=100, label='Threshold')
ax.legend()
plt.title("Reconstruction error for different classes")
plt.ylabel("Reconstruction error")
plt.xlabel("Data point index")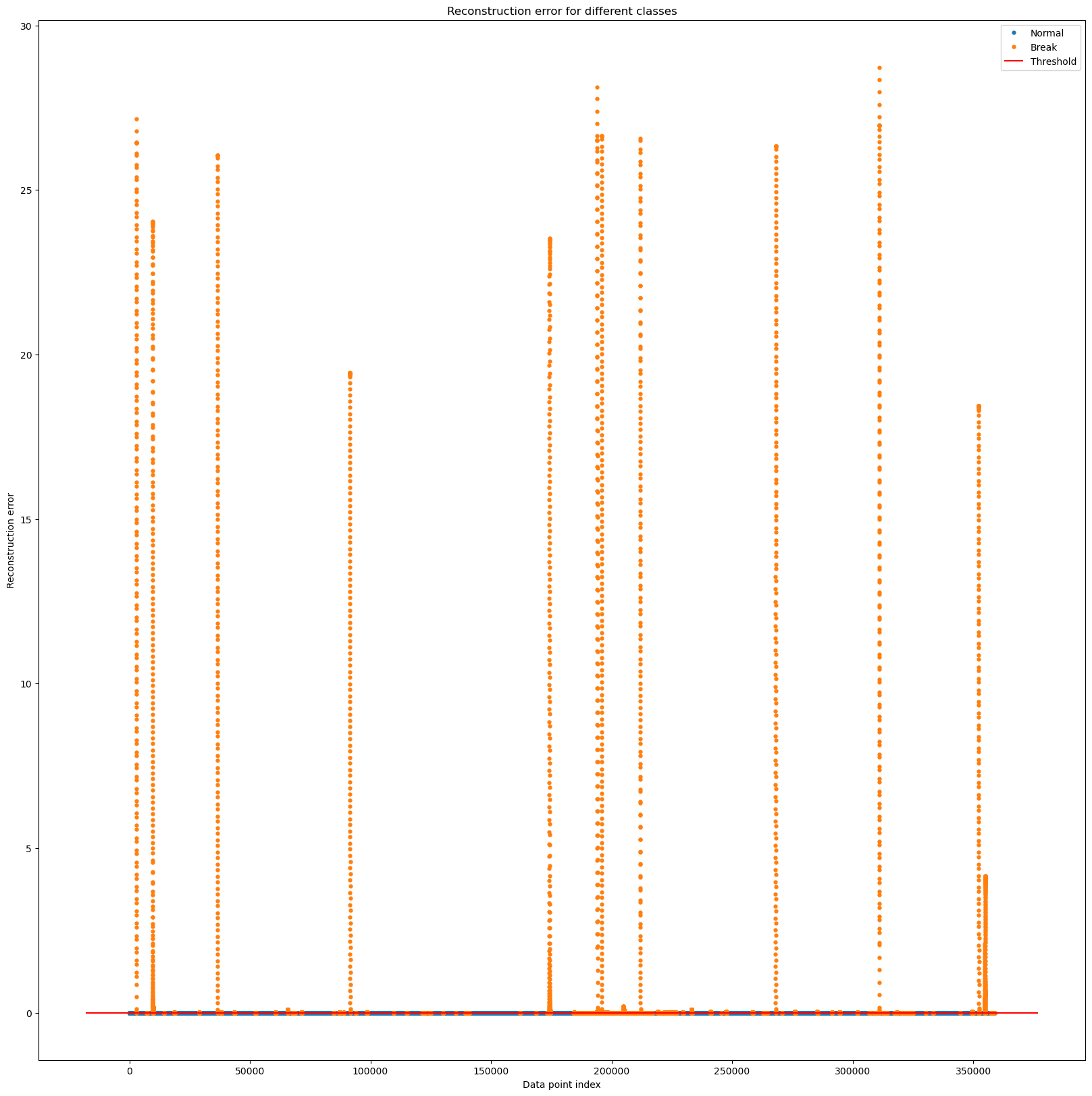
728x90
반응형
'👩💻 인공지능 (ML & DL) > Serial Data' 카테고리의 다른 글
| [DACON] 동서발전 태양광 발전량 예측 AI 경진대회 (0) | 2022.09.14 |
|---|---|
| 시계열 모델 ARIMA 2 (자기회귀 집적 이동 평균) (0) | 2022.09.08 |
| [이상 탐지] ML for Time Series & windows (0) | 2022.09.07 |
| [DACON] HAICon2020 산업제어시스템 보안위협 탐지 AI & LSTM (0) | 2022.09.07 |
| ADTK (Anomaly detection toolkit) 시계열 이상탐지 오픈소스 (0) | 2022.09.06 |
Comments




